mailto:werner@yellowcouch.org

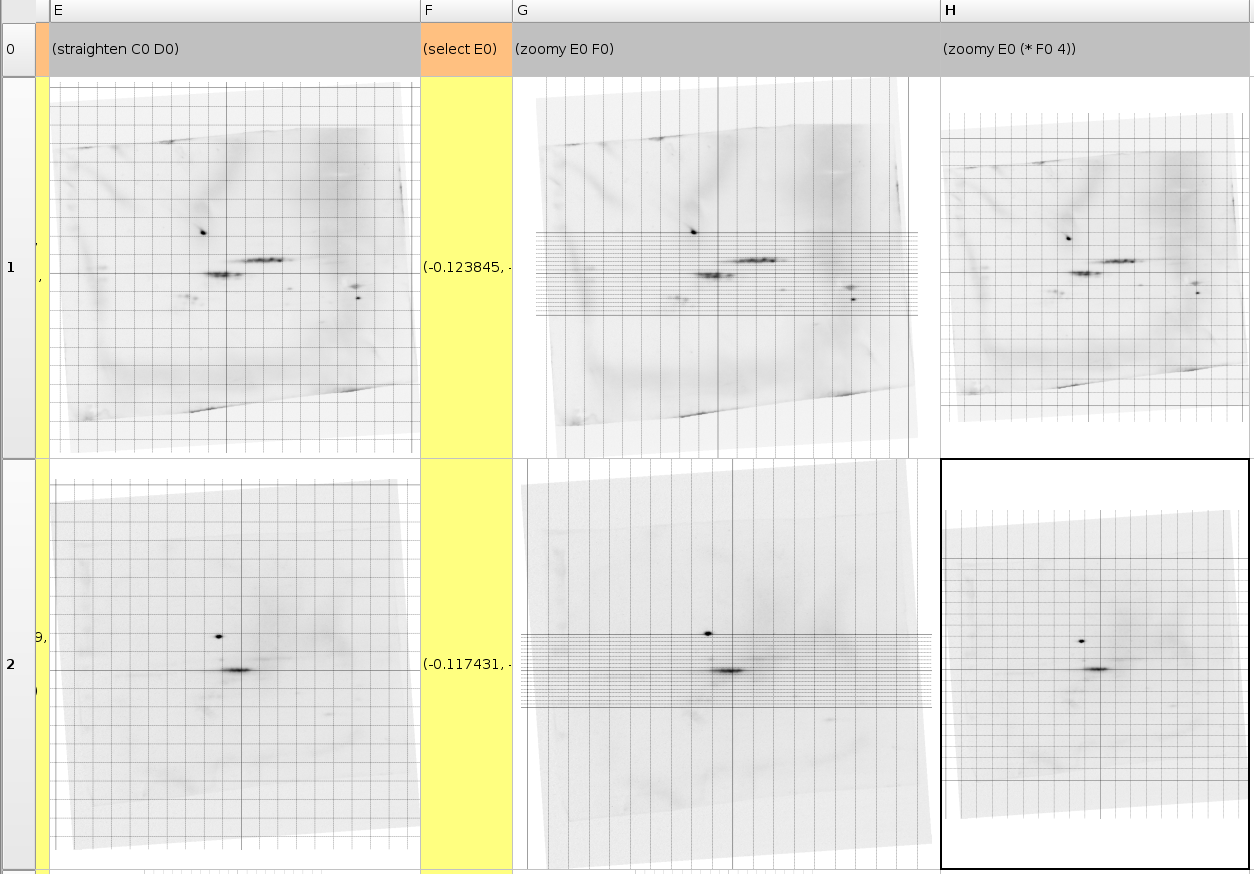
(select E0)After the appropriate image rotation and centering has been performed we can determine the proper scaling on both axes: the pI axis and the mass axis. Since the pI axe is in this example created using precast strips we can assume that there are no scaling problems. The time based mass separation on the other hand has a whole other error. Therefore it is important to establish the distance between our chosen center and another protein we believe is accurately measured. In our case we assume that the p63 spot can be used as a scaling means. Practically we need to mark the p63 spot on all the gels using a select command and then use the zoom operation to change the coordinate system such that the p63 spot its height along the y-axis becomes 1.0. Of course, such a coordinate system is not entirely as we want it, therefore it could be useful to scale the axis with a factor 4 such that the y-position of the p63 spot becomes 0.25. This can be done with (zoom E0 (* F0 4)).
(zoomy E0 F0)
(zoomy E0 (* F0 4))
| http://werner.yellowcouch.org/ mailto:werner@yellowcouch.org |  |